Using the InteractiveMolecule widget#
For this example we need to install RDKit. Note that you will need conda to install RDKit.
!conda install -c conda-forge rdkit
Now we can import the trident_chemwidgets and the pandas lib to import our csv dataset.
[1]:
import trident_chemwidgets as tcw
import pandas as pd
from rdkit import Chem
---------------------------------------------------------------------------
ModuleNotFoundError Traceback (most recent call last)
Input In [1], in <cell line: 3>()
1 import trident_chemwidgets as tcw
2 import pandas as pd
----> 3 from rdkit import Chem
ModuleNotFoundError: No module named 'rdkit'
Now we can create a small function to featurize our molecules with basic information per atom.
IMPORTANT: the order of the data rows in the pandas DataFrame or dict must match the standard ordering of atoms as returned by the RDKit ``.GetAtoms()`` function. You can generate this data any way you see fit (e.g. calculated values from RDKit as in the function below or attention values from a Graph Attention Network. The only constraint is the atom ordering. If you are using RDKit-based featurizers like those from DeepChem, this standard ordering should already be the default. Take care when using cutom featurizers.
[2]:
def featurize_mol(smiles):
# Init feature dict
feature_dict = {
'Chiral Tag': [],
'Formal Charge': [],
'Mass': [],
'Total Hs': [],
'Total Valence': []
}
# Get atoms from SMILES
atoms = Chem.MolFromSmiles(smiles).GetAtoms()
# Use RDKit to get all the atom properties
for atom in atoms:
feature_dict['Chiral Tag'].append(atom.GetChiralTag())
feature_dict['Formal Charge'].append(atom.GetFormalCharge())
feature_dict['Mass'].append(atom.GetMass())
feature_dict['Total Hs'].append(atom.GetTotalNumHs())
feature_dict['Total Valence'].append(atom.GetTotalValence())
return pd.DataFrame.from_dict(feature_dict)
Here we’ll be exploring the atom features from the ibuprofen molecule, smiles string CC(C)CC1=CC=C(C=C1)C(C)C(=O)O. We’ll use the function we defined above to get some data at the atom level.
[3]:
atom_data = featurize_mol('CC(C)CC1=CC=C(C=C1)C(C)C(=O)O')
atom_data.head()
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Input In [3], in <cell line: 1>()
----> 1 atom_data = featurize_mol('CC(C)CC1=CC=C(C=C1)C(C)C(=O)O')
2 atom_data.head()
Input In [2], in featurize_mol(smiles)
3 feature_dict = {
4 'Chiral Tag': [],
5 'Formal Charge': [],
(...)
8 'Total Valence': []
9 }
11 # Get atoms from SMILES
---> 12 atoms = Chem.MolFromSmiles(smiles).GetAtoms()
14 # Use RDKit to get all the atom properties
15 for atom in atoms:
NameError: name 'Chem' is not defined
Now we can use the InteractiveMolecule widget to explore the data attached to each atom.
[4]:
w = tcw.InteractiveMolecule('CC(C)CC1=CC=C(C=C1)C(C)C(=O)O', data=atom_data)
# w # Uncomment this line to run locally
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Input In [4], in <cell line: 1>()
----> 1 w = tcw.InteractiveMolecule('CC(C)CC1=CC=C(C=C1)C(C)C(=O)O', data=atom_data)
NameError: name 'atom_data' is not defined
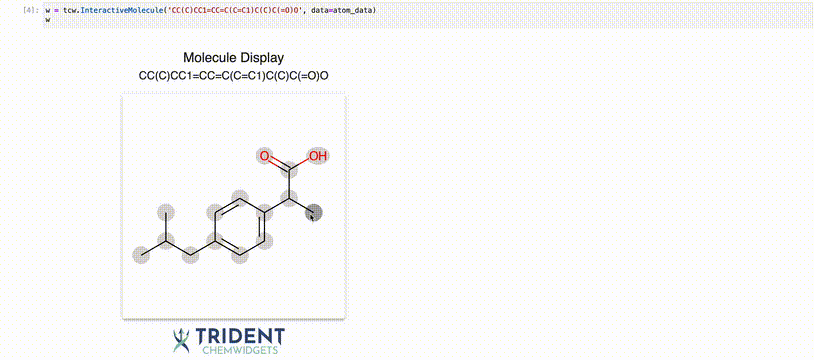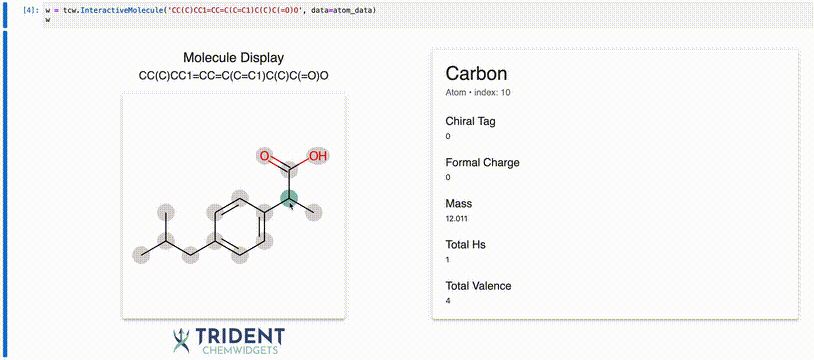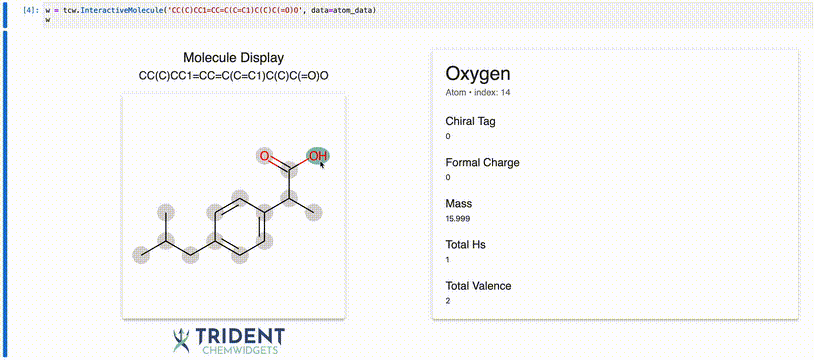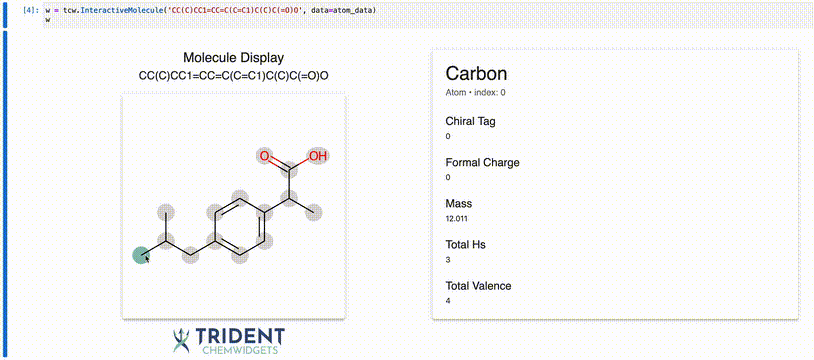
The value of the widget will match what you typed into the input.
[5]:
w.smiles
---------------------------------------------------------------------------
NameError Traceback (most recent call last)
Input In [5], in <cell line: 1>()
----> 1 w.smiles
NameError: name 'w' is not defined